Previous: smoothing Up: HEALPix/F90 facilities Next: ud_grade Top: Main Page
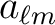 coefficients and/or power spectra.
Total operation count scales as
coefficients and/or power spectra.
Total operation count scales as
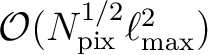 where
Npix is the total number of pixels and
where
Npix is the total number of pixels and
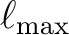 is the limiting spherical harmonics order.
The map resolution, Gaussian beam FWHM,
and random seed for the simulation can be selected by the user.
Spherical harmonics are either generated using the recurrence relations
during the execution of spectral synthesis, or precomputed and read in
before the synthesis is executed. The latter is no longer recommended since
it provides no acceleration since the introduction of optimized algorithms
in HEALPix v2.20.
is the limiting spherical harmonics order.
The map resolution, Gaussian beam FWHM,
and random seed for the simulation can be selected by the user.
Spherical harmonics are either generated using the recurrence relations
during the execution of spectral synthesis, or precomputed and read in
before the synthesis is executed. The latter is no longer recommended since
it provides no acceleration since the introduction of optimized algorithms
in HEALPix v2.20.
% synfast [options] [parameter_file]
 generated can be output.
generated can be output.
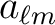 used
for the simulation (default= `')
used
for the simulation (default= `')
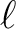 value
to be used in the simulation. WARNING:
value
to be used in the simulation. WARNING:
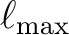 can not exceed
the value
can not exceed
the value 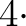 nsmax, because the coefficients of the average Fourier
pixel window functions
are precomputed and provided up to this limit.
(default= 64)
nsmax, because the coefficients of the average Fourier
pixel window functions
are precomputed and provided up to this limit.
(default= 64)
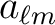 s from the power spectrum.
(default= -1)
s from the power spectrum.
(default= -1)
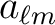 s for constrained realisations.
(default= `'). If apply_windows is false
those
s for constrained realisations.
(default= `'). If apply_windows is false
those 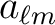 s are used as they are, without being multiplied
by the beam or pixel window function (with the assumption that they already have the
correct window functions). If apply_windows is true, the beam and
pixel window functions chosen above are applied to the constraining
s are used as they are, without being multiplied
by the beam or pixel window function (with the assumption that they already have the
correct window functions). If apply_windows is true, the beam and
pixel window functions chosen above are applied to the constraining 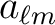 (with the
assumption that those are free of beam and pixel window function). The code
does not check the validity of these asumptions; if none is true, use the
alteralm facility to modify or remove
the window functions contained in the constraining
(with the
assumption that those are free of beam and pixel window function). The code
does not check the validity of these asumptions; if none is true, use the
alteralm facility to modify or remove
the window functions contained in the constraining 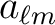 .
.
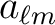 read from
almsfile are
treated with respect to window functions; see above for details.
y, yes, t, true, .true. and 1 are considered as true, while n, no, f,
false, .false. and 0 are considered as false, (default = .false.).
read from
almsfile are
treated with respect to window functions; see above for details.
y, yes, t, true, .true. and 1 are considered as true, while n, no, f,
false, .false. and 0 are considered as false, (default = .false.).
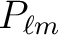 .
(default= `')
.
(default= `')
Synfast reads the power spectrum from a file in ascii FITS format. This can contain either just the temperature power spectrums or temperature and polarisation power spectra:
,
,
and
(see Note 1, below). If simul_type = 2 synfast generates Q and U maps as well as the temperature map. The output map(s) is (are) saved in a FITS file. The
s are used up to the specified
, which can not exceed 4 x nsmax. If simul_type = 3 or 4 the first derivatives of the temperature field or the first and second derivatives respectively are output as well as the temperature itself: T(p),
,
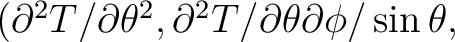
. If simul_type = 5 or 6 the first derivatives of the (T,Q,U) fields or the first and second derivatives respectively are output as well as the field themself: T(p), Q(p), U(p),
,
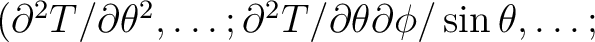
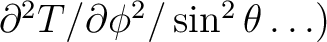
The random sequence seed for generation offrom the power spectrum should be non-zero integer. If 0 is provided, a seed is generated randomly by the code, based on the current date and time. The map can be convolved with a gaussian beam for which a beamsize can be specified, or for an arbitrary circular beam for which the Legendre transform is provided. The map is automatically convolved with a pixel window function. These are stored in FITS files in the healpix/data directory. If synfast is not run in a directory which has these files, or from a directory which can reach these files by a `../data/' or `./data/' specification, the system variable HEALPIX is used to locate the main HEALPix directory and its data subdirectory is scanned. Failing this, the location of these files must be specified (using winfiledir). In the interactive mode this is requested only when necessary (see Notes on default directories ).
If some of thein the simulations are constrained eg. from observations, a FITS file with these
can be read. This FITS file contains the
for certain
and m values and also the standard deviation for these
. The sky realisation which synfast produces will be statistically consistent with the constraining
.
The code can also be used to generate a set ofmatching the input power spectra, beam size and pixel size with or without actually synthesizing the map. Those
can be used as an input (constraining
) to another synfast run.
Spherical harmonics values in the synthesis are obtained from a recurrence on associated Legendre polynomials. This recurrence consumed most of the CPU time used by synfast up to version 2.15. We have therefore included an option to load precomputed values for the
from a file generated by the HEALPix facility plmgen. Since the introduction of accelerated spherical harmonic transforms in HEALPix v2.20, this feature is obsolete and should no longer be used.
synfast will issue a warning if the input FITS file for the power spectrum does not contain the keyword POLNORM. This keyword indicates that the convention used for polarization is consistent with CMBFAST (and consistent with HEALPix 1.2). See the HEALPix Primer for details on the polarization convention and the interface with CMBFAST. If the keyword is not found, no attempt will be made to renormalize the power spectrum. If the keyword is present, it will be inherited by the simulated map.
Note 1: to allow the generation of maps (and
) with
and/or
, see the subroutine create_alm.
| Dataset | Description |
|---|---|
| /data/pixel_window_nxxxx.fits | Files containing pixel windows for various nsmax. |
 window function used in synfast
window function used in synfast
 s (almsfile file).
s (almsfile file).
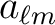 and
and  s to be read by synfast.
s to be read by synfast.
| synfast |
Synfast runs in interactive mode, self-explanatory.
| synfast filename |
When 'filename' is present, synfast enters the non-interactive mode and parses its inputs from the file 'filename'. This has the following structure: the first entry is a qualifier which announces to the parser which input immediately follows. If this input is omitted in the input file, the parser assumes the default value. If the equality sign is omitted, then the parser ignores the entry. In this way comments may also be included in the file. In this example, the file contains the following qualifiers:
simul_type= 1
nsmax= 32
nlmax= 64
iseed= -1
fwhm_arcmin= 420.0
infile= cl.fits
outfile= map.fits
Synfast reads in the
power spectrum in 'cl.fits' up to
, and produces the (RING ordered) map 'map.fits' which has Nside=32. The map is convolved with a beam of FWHM 420.0 arcminutes. The iseed=-1 sets the random seed for the realisation. A different iseed would have given a different realisation from the same power spectrum.
Since
outfile_alms
almsfile
apply_windows
plmfile
beam_file
windowfile
were omitted, they take their default values (empty strings). This means that no file for constrained realisation or precomputed Legendre polynomials are read, thegenerated in the process are not output, and synfast attempts to find the pixel window files in the default directories (see Notes on default files and directories).
- ■ Initial release (HEALPix 0.90)
- ■ Optional non-interactive operation. Proper FITS file support. Improved reccurence algorithm for
which can compute to higher
values. Improved pixel windows averaged over actual HEALPix pixels. New functionality: constrained realisations, precomputed
. (HEALPix 1.00)
- ■ New functionality: constrained realisations and pixel windows are now available for polarization as well. Arbitrary circular beams can be used. New parser (HEALPix 1.20)
- ■ New functionnality: the generated
can be output, and the map synthesis itself can be skipped. First and second derivatives of the temperature field can be produced on demand.
- ■ New functionnality: First and second derivatives of the Q and U Stokes field can be produced on demand.
- ■ Bug correction: corrected numerical errors on derivatives
,
,
, for X=Q,U. See this appendix for details. (HEALPix 2.14)
| Message | Severity | Text |
|---|---|---|
| can not allocate memory for array xxx | Fatal | You do not have sufficient system resources to run this facility at the map resolution you required. Try a lower map resolution. |
|
this is not a binary table |
the fitsfile you have specified is not of the proper format | |
| there are undefined values in the table! | the fitsfile you have specified is not of the proper format | |
| the header in xxx is too long | the fitsfile you have specified is not of the proper format | |
| XXX-keyword not found | the fitsfile you have specified is not of the proper format | |
| found xxx in the file, expected:yyyy | the specified fitsfile does not contain the proper amount of data. | |
Version 3.83, 2024-11-13