Previous: median_filter Up: HEALPix/F90 facilities Next: process_mask Top: Main Page
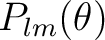 (and, if requested, of the tensor spherical harmonics)
for faster execution of the HEALPix map analysis/synthesis.
The map resolution parameter, nsmax,
and the maximum value of the spherical harmonic order
(and, if requested, of the tensor spherical harmonics)
for faster execution of the HEALPix map analysis/synthesis.
The map resolution parameter, nsmax,
and the maximum value of the spherical harmonic order 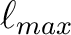 must be specified.
must be specified.
% plmgen
 value for the execution.
(default= 64)
value for the execution.
(default= 64)
The recursion of Legendre polynomials and tensor harmonics during the analysis and synthesis of HEALPix maps can be time consuming. Especially when repetitive applications are desired there is no need to compute the recursions every time. For such applications the values ofcan be precomputed with plmgen and stored in a file. When using synfast or anafast this file can be read in to shorten the analysis/synthesis execution time.
The memory (and disc) consumption of plmgen isbytes, with
and Np is either 1 or 3, depending whether tensor harmonics are computed.
Currently an extra limitationalso applies, corresponding to, eg, lmax
for nsmax =1024.
| Dataset | Description |
|---|---|
| None required | |
| plmgen |
plmgen runs in interactive mode, self-explanatory.
| plmgen filename |
When `filename' is present, plmgen enters the non-interactive mode and parses its inputs from the file `filename'. This has the following structure: the first entry is a qualifier which announces to the parser which input immediately follows. If this input is omitted in the input file, the parser assumes the default value. If the equality sign is omitted, then the parser ignores the entry. In this way comments may also be included in the file. In this example, the file contains the following qualifiers:
simul_type= 1
nsmax= 32
nlmax= 86
outfile= plm.fits
Creates a binary FITS file called 'plm.fits' containing Legendre polynomials up to
and m values of 86 for a nsmax=32 map. Legendre polynomials for all
and m values for each angle
corresponding to all of the HEALPix pixel center rings will be created.
- ■ Initial release HEALPix 1.00
| Message | Severity | Text |
|---|---|---|
| can not allocate memory for array xxx | Fatal | You do not have sufficient system resources to run this facility at the map resolution you required. Try a lower map resolution. |
Error: these values of Nside and l_max  are too large are too large  |
Fatal | You are exceeding the limitation on Nside and l_max. Try a lower l_max. |
Version 3.83, 2024-11-13