Previous: coordsys2euler_zyz Up: HEALPix Fortran90 Subroutines Overview Next: del_card Top: Main Page
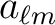 for a temperature (and
polarisation) power spectrum read from an input FITS
file. The
for a temperature (and
polarisation) power spectrum read from an input FITS
file. The
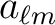 are gaussian distributed with a zero mean, and their
amplitude is multiplied with the
are gaussian distributed with a zero mean, and their
amplitude is multiplied with the 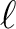 -space window function of a gaussian
beam characterized by its FWHM or an arbitrary circular beam
and a pixel window read from an external file.
-space window function of a gaussian
beam characterized by its FWHM or an arbitrary circular beam
and a pixel window read from an external file.
Arguments appearing in italic are optional.call create_alm*( nsmax, nlmax, nmmax, polar, filename, rng_handle, fwhm_arcmin, alm_TGC, header[, windowfile, units, beam_file] )
| name & dimensionality | kind | in/out | description |
|---|---|---|---|
| nsmax | I4B | IN |
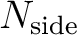 of the map to be synthetized from the of the map to be synthetized from the
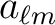 created by this routine.
created by this routine. |
| nlmax | I4B | IN | maximum 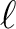 value to be considered (MAX= value to be considered (MAX=
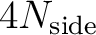 if windowfile is provided).
if windowfile is provided). |
| nmmax | I4B | IN | maximum 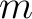 value for the value for the
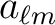 . . |
| polar | I4B | IN | if set to 0, only Temperature (scalar)
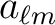 are
generated using TT spectrum. If set to 1, 'conventional' polarization is added, based on EE, BB and TE spectra. If set to 2, and if
the relevant information is in filename, polarization is generated
assuming non-zero correlation of Curl (B) modes with Temperature (T) and Gradient
(E) modes (TB and EB cross-spectra). Note that the synfast facility calls create_alm* with polar=0 or polar=1 are
generated using TT spectrum. If set to 1, 'conventional' polarization is added, based on EE, BB and TE spectra. If set to 2, and if
the relevant information is in filename, polarization is generated
assuming non-zero correlation of Curl (B) modes with Temperature (T) and Gradient
(E) modes (TB and EB cross-spectra). Note that the synfast facility calls create_alm* with polar=0 or polar=1 |
| filename(LEN=filenamelen) | CHR | IN | name of FITS file containing power spectra in the order TT, [EE, BB, TE, [TB, EB]] (terms in brackets are optional, see polar) |
| rng_handle | planck_rng | INOUT | structure containing information necessary to continue a random sequence initiated previously with the subroutine rand_init. Consecutive calls to create_alm* can be made after a single invocation to rand_init. |
| fwhm_arcmin | SP/ DP | IN | FWHM size of the gaussian beam in arcminutes. |
| alm_TGC(1:p,0:nlmax,0:nmmax) | SPC/ DPC | OUT | complex
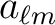 values
generated from the power spectrum in the FITS file. The first index here runs
form 1:1 for temperature only, and 1:3 for polarisation. In the latter case,
1=T, 2=E, 3=B. values
generated from the power spectrum in the FITS file. The first index here runs
form 1:1 for temperature only, and 1:3 for polarisation. In the latter case,
1=T, 2=E, 3=B. |
| name & dimensionality | kind | in/out | description |
|---|---|---|---|
| header(LEN=80),dimension(60) | CHR | OUT | part of header which
will be included in the FITS-file containing the
map synthesised from the
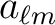 which create_alm generates. which create_alm generates. |
| windowfile(LEN=filenamelen) | CHR | IN | full filename specification
of the FITS file with the pixel window function (defined for
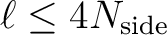 ) ) |
| units(LEN=80),dimension(1:) | CHR | OUT | physical units of the created
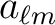 (square-root of the input power spectrum units). (square-root of the input power spectrum units). |
| beam_file(LEN=filenamelen) | CHR | IN | name of the file containing
the (non necessarily gaussian) window function 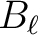 of a circular beam. If present, it will override
the argument fwhm_arcmin. of a circular beam. If present, it will override
the argument fwhm_arcmin. |
Creates scalar and tensorfrom the power spectrum given in the file `cl.fits'. The map to be created from these
is assumed to have
.
s from the power spectrum are used up to an
value of 128. Corresponding
values up to l=128 and m=128 are created as gaussian distributed complex numbers. Their are drawn from a sequence of pseudo-random numbers initiated with a seed of -1. The produced
are convolved with a gaussian beam of FWHM 5 arcminutes and a pixel window read from 'data/pixel_window_n0064.fits'. It is assumed that after the return from this routine, a map is generated from the created
. For this purpose, header is updated with FITS format information describing the origin and history of these
.
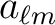 units
units
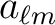 created by create_alm* to a HEALPix map.
created by create_alm* to a HEALPix map.
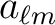 in a FITS file.
in a FITS file.
Version 3.83, 2024-11-13