Previous: read_fits_s Up: HEALPix/IDL subroutines Next: remove_dipole Top: Main Page
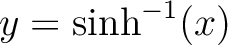 3 is
the number of extensions read
3 is
the number of extensions read
read_tqu reads out Stokes parameters (T,Q,U) maps for the whole sky into a FITS file. It is also possible to read the error per pixel for each map and the correlation between fields, as subsequent extensions of the same FITS file (see qualifiers above). Therefore the file may have up to three extensions with three maps in each. Extensions can be written together or one by one (in their physical order) using the Extension option.
For more information on the FITS file format supported in HEALPix, including the one implemented in read_tqu , see https://healpix.sourceforge.io/data/examples/healpix_fits_specs.pdf.
| read_tqu, 'map_polarization.fits', TQU, xhdr=xhdr |
Reads into TQU the polarization maps contained in the FITS file 'map_polarization.fits'. The variable xhdr will contain the extension(s) header.
Version 3.83, 2024-11-13