Previous: fits2alm Up: HEALPix/IDL subroutines Next: gaussbeam Top: Main Page
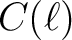 ) or spherical harmonics (
) or spherical harmonics (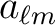 ) coefficients, and returns
the corresponding power spectrum (
) coefficients, and returns
the corresponding power spectrum (
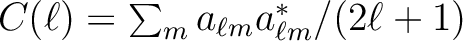 ). Reads primary and extension headers if
required. The facility is intended to enable the user to read the
output from the HEALPix facility anafast.
). Reads primary and extension headers if
required. The facility is intended to enable the user to read the
output from the HEALPix facility anafast.
 coefficients read or computed from the
file. The output dimension depends on the contents of the file.
This has dimension either (lmax+1,9) given in the sequence T E B
TxE TxB ExB ExT BxT BxE or
(lmax+1,6) given in the sequence T E B
TxE TxB ExB or (lmax+1,4) for T E B TxE or (lmax+1) for T
alone.
coefficients read or computed from the
file. The output dimension depends on the contents of the file.
This has dimension either (lmax+1,9) given in the sequence T E B
TxE TxB ExB ExT BxT BxE or
(lmax+1,6) given in the sequence T E B
TxE TxB ExB or (lmax+1,4) for T E B TxE or (lmax+1) for T
alone.
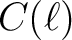 power spectra or
power spectra or 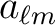 coefficients. In either
cases,
coefficients. In either
cases, 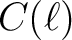 is returned. If fitsfile is not set, then
/PLANCK1,
/PLANCK2,
/PLANCK3,
/WMAP1,
/WMAP5 or
/WMAP7
must be set.
is returned. If fitsfile is not set, then
/PLANCK1,
/PLANCK2,
/PLANCK3,
/WMAP1,
/WMAP5 or
/WMAP7
must be set.
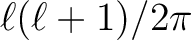 which is often
applied to
which is often
applied to 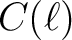 to flatten it for plotting purposes
to flatten it for plotting purposes
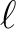 for which the power spectra are provided. They are either
for which the power spectra are provided. They are either
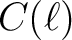 read from the file are plotted
read from the file are plotted
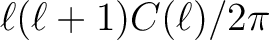 are plotted
are plotted
fits2cl reads the power spectrum coefficients from a FITS file containing an ascii table extension. Descriptive headers conforming to the FITS convention can also be read from the input file.
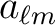 coefficients
coefficients
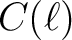 files
that can be read by fits2cl.
files
that can be read by fits2cl.
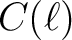 files that can be read by fits2cl.
files that can be read by fits2cl.
| fits2cl, pwrsp, '$HEALPIX/test/cl.fits', $ |
| HDR=hdr, XHDR=xhdr, MULTI=l, LLFACT=fll |
| plot, l, powrsp[*,0]*fll |
fits2cl reads a power spectrumfrom the input FITS file $HEALPIX/test/cl.fits into the variable pwrsp, with optional headers passed by the string variables hdr and xhdr. The multipoles
and factors
are read into l and fll respectively.
vs
is then plotted.
Version 3.83, 2024-11-13