Previous: change_polcconv Up: HEALPix/IDL subroutines Next: convert_oldhpx2cmbfast Top: Main Page
cl2fits writes the input power spectrum coefficients into a FITS file containing an ascii table extension. Optional headers conforming to the FITS convention can also be written to the output file. All required FITS header keywords (like SIMPLE, BITPIX, ...) are automatically generated by the routine and should NOT be duplicated in the optional header inputs (they would be ignored anyway). The one/four/six/nine column(s) are automatically named TEMPERATURE, GRAD, CURL, TG, TC, GC, GT, CT and CG respectively. If the power spectrum is provided in a double precision array, the output format will automatically feature more decimal places. The current implementation is much faster than the one available in Healpix 1.10 thanks to replacing an internal loop by vector operations.
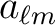 coefficients
coefficients
| cl2fits, pwrsp, 'spectrum.fits', HDR = hdr, XHDR = xhdr |
cl2fits writes the power spectrum stored in the variable pwrsp to the output FITS file spectrum.fits with optional headers passed by the string variables hdr and xhdr.
Version 3.83, 2024-11-13