Previous: read_fits_partial Up: HEALPix Fortran90 Subroutines Overview Next: real_fft Top: Main Page
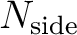 , number of
datasets (maps) and maximum multipole
, number of
datasets (maps) and maximum multipole 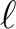 (order) and
(order) and 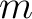 (degree) value
for the file. If a keyword is not found in the FITS file, a value of -1 is
returned instead. The file could eg. be a HEALPix map, or a set of
(degree) value
for the file. If a keyword is not found in the FITS file, a value of -1 is
returned instead. The file could eg. be a HEALPix map, or a set of
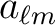 or precomputed
or precomputed
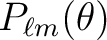
call read_par( filename, nside, lmax, tfields[, mmax] )
Checks theand maximum
value used for the precomputed
that are stored in the file `plm_128p.fits'. If the file also contains tensor harmonics, nhar is returned with the value 3, otherwise it is 1.
Version 3.83, 2024-11-13