Previous: euler_matrix_new Up: HEALPix/IDL subroutines Next: fits2cl Top: Main Page
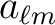 (and optional errors) and their index. Reads
header information if required. The facility is intended to enable
the user to read the output from the HEALPix facilities anafast and synfast.
(and optional errors) and their index. Reads
header information if required. The facility is intended to enable
the user to read the output from the HEALPix facilities anafast and synfast.
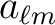 coefficients (and errors if required). The
index i is related to
coefficients (and errors if required). The
index i is related to 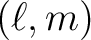 by the relation
by the relation 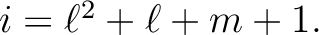
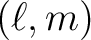 indices
indices
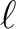 multipole to be output
multipole to be output
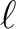 multipole to be output. If LMIN (resp. LMAX) is below (above) the range of l's present in the file,
it will be silently ignored
multipole to be output. If LMIN (resp. LMAX) is below (above) the range of l's present in the file,
it will be silently ignored
fits2alm reads binary table extension(s) which contain thecoefficients (and associated errors if present) from a FITS file. FITS headers can also optionally be read from the input file.
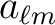 coefficients into a FITS file.
coefficients into a FITS file.
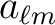 into 2D a(l,m) tables and back
into 2D a(l,m) tables and back
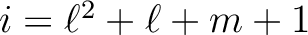 returned by fits2alm into
returned by fits2alm into 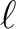 and m
and m
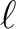 , m) vectors into
, m) vectors into
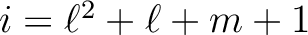
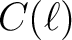 power spectra from a file containing
power spectra from a file containing 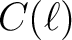 or
or 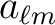 coefficients
coefficients
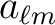 coefficients file to be read by fits2alm.
coefficients file to be read by fits2alm.
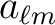 coefficients file to be read by fits2alm.
coefficients file to be read by fits2alm.
| fits2alm, index, alm, 'alm.fits', HDR = hdr, XHDR = xhdr |
fits2alm reads from the input FITS file alm.fits thecoefficients into the variable alm with optional headers passed by the string variables hdr and xhdr. Upon return index will contain the value of
for each
found in the file.
Version 3.83, 2024-11-13