Previous: mpi_cleanup_alm_tools Up: HEALPix Fortran90 Subroutines Overview Next: mpi_map2alm* Top: Main Page
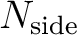 and
and
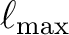 , since each processor only needs a fraction
, since each processor only needs a fraction
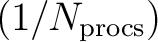 of the complete table. This feature is
controlled by the “precompute_plms” parameter. In general, the CPU
time can be expected to decrease by roughly 50% using pre-computed
Legendre polynomials for temperature calculations, and by about 30%
for polarization calculations.
of the complete table. This feature is
controlled by the “precompute_plms” parameter. In general, the CPU
time can be expected to decrease by roughly 50% using pre-computed
Legendre polynomials for temperature calculations, and by about 30%
for polarization calculations.
call mpi_initialize_alm_tools( comm, [nsmax], [nlmax], [nmmax], [zbounds], [polarization], [precompute_plms], [w8ring] )
This example 1) initializes the mpi_alm_tools module (i.e., allocates internal arrays and defines required parameters), 2) executes a parallel map2alm operation, and 3) frees the previously allocated memory.
Version 3.83, 2024-11-13